Help
-
DiseaseMeth version 3.0
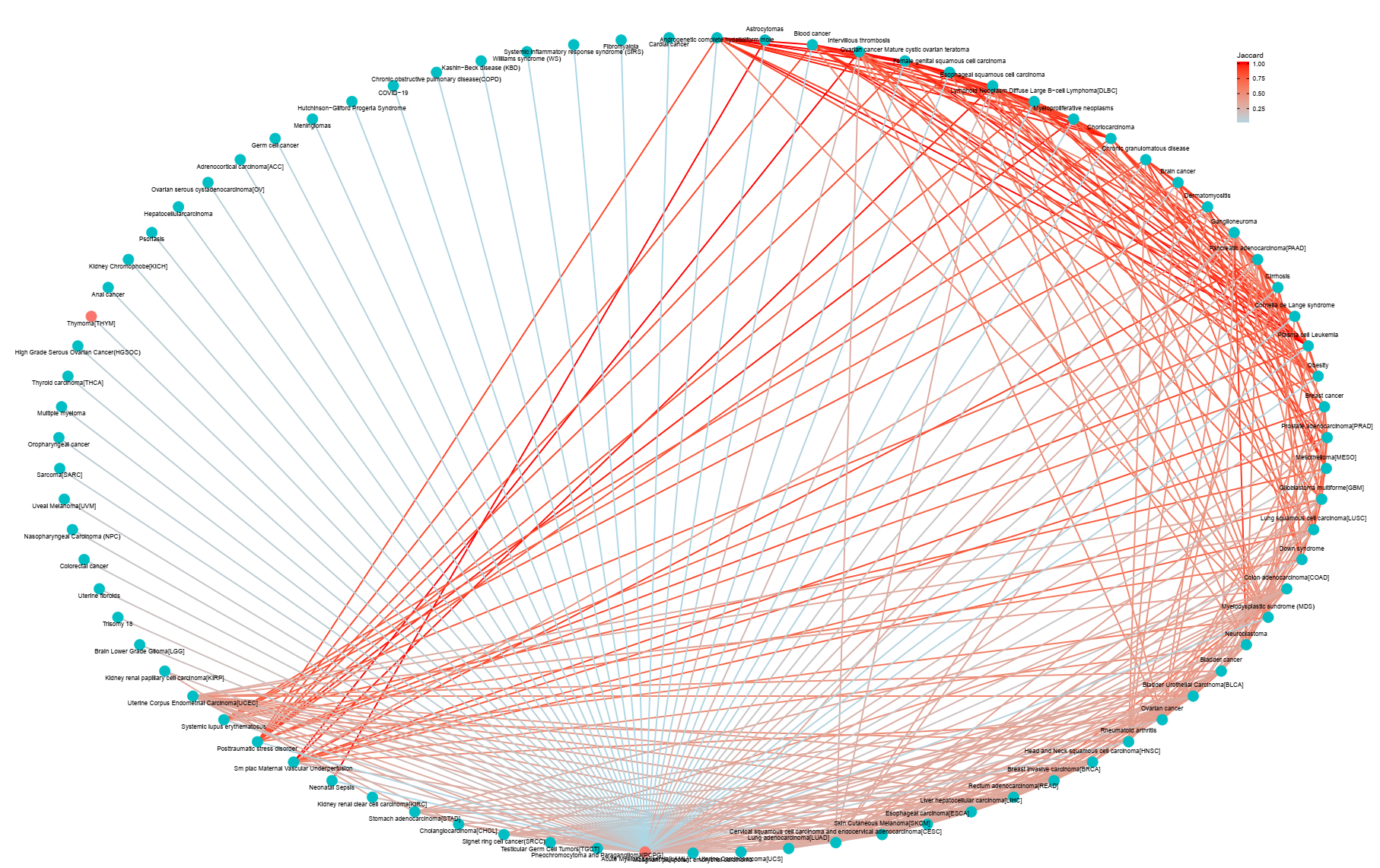
The human disease DNA methylation database, DiseaseMeth version 3.0 is a web based resource focused on the aberrant methylomes of human diseases. Until recently, bulks of large-scale data are avaible and are increasingly grown, from which more information can be mined to gain further information towards human diseases. Our mission is to provide a curated set of methylation information datasets and tools in the human genome, to support and promote research in this area. Especially, based on the DMGs, we calculated correlations between genes in 162 diseases and provided the visualization results by network. Key genes can be characterized by DNA methylation correlation networks.
-
Architecture Design
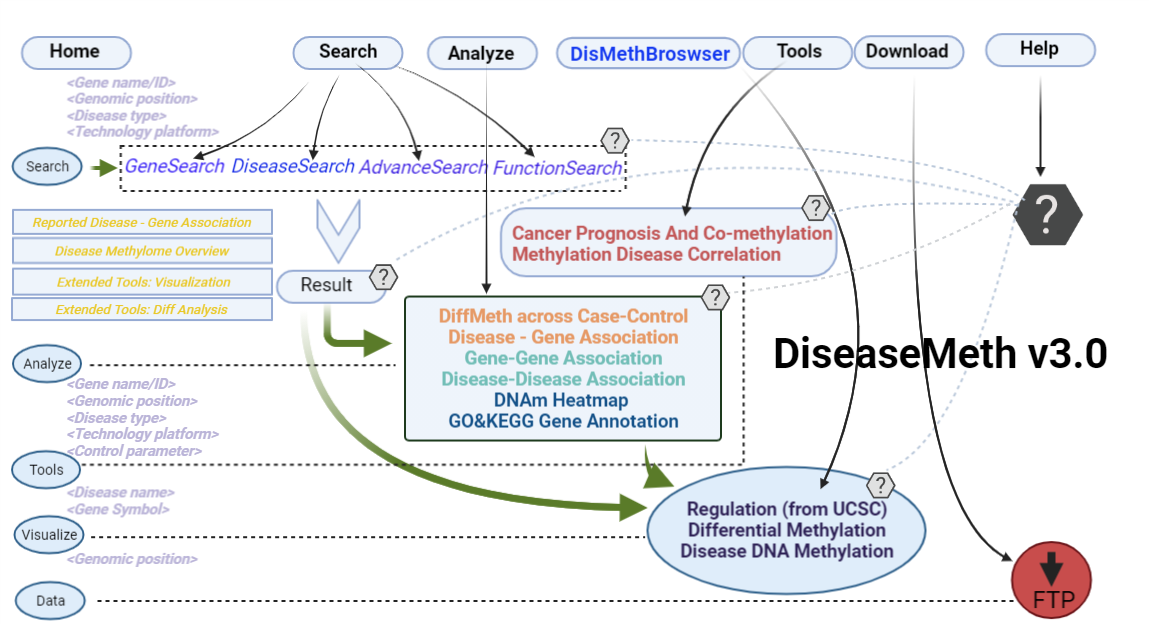
-
the blue trace
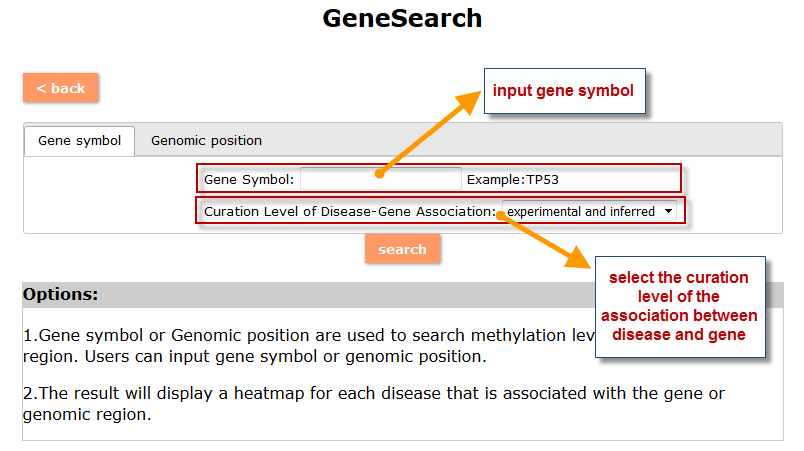
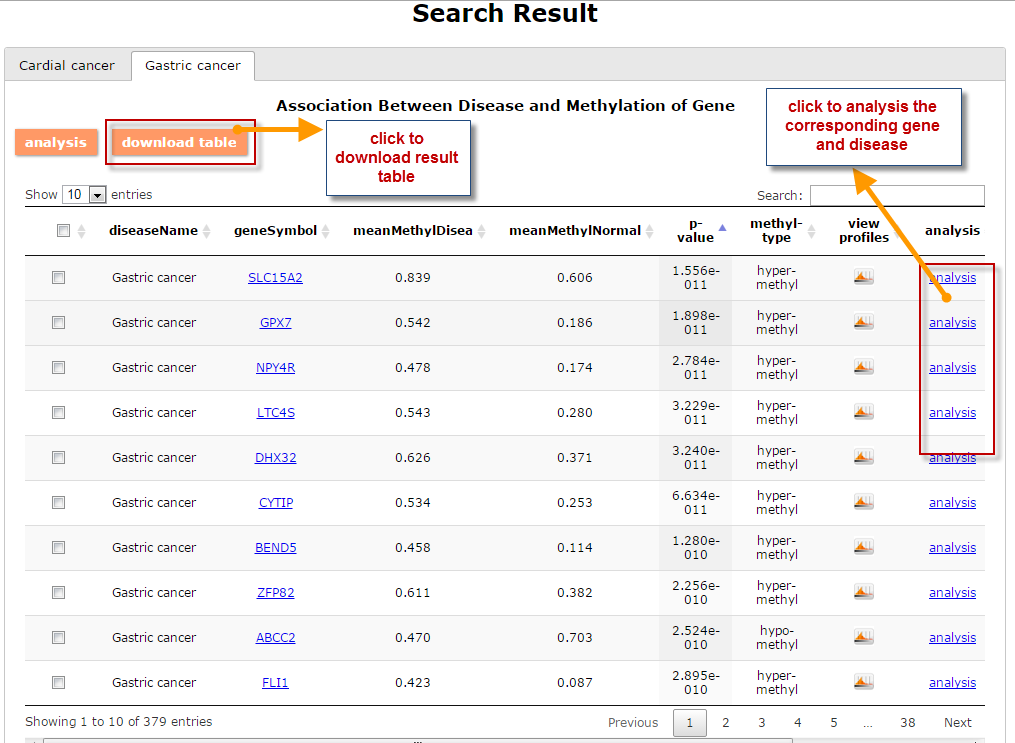
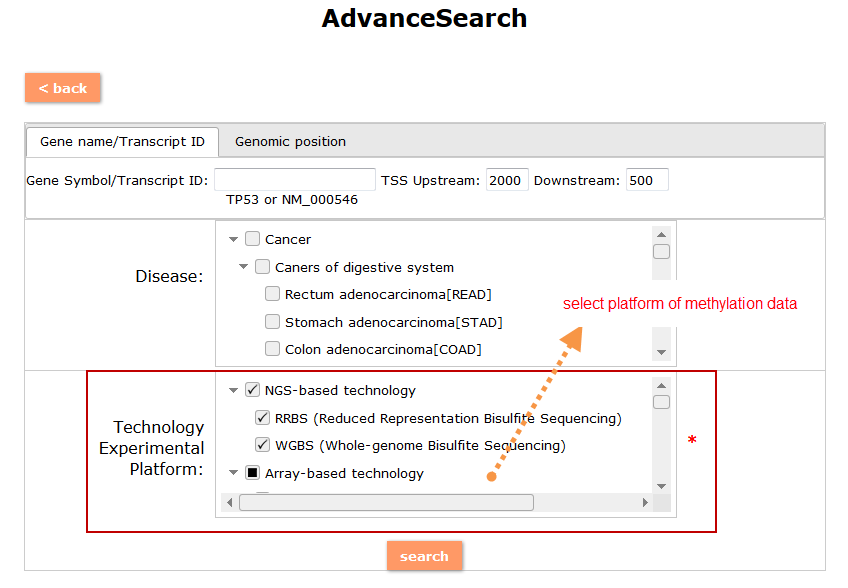
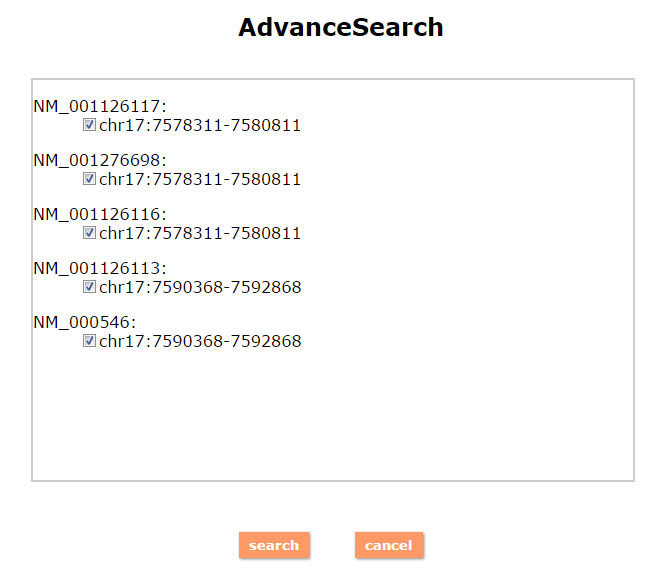
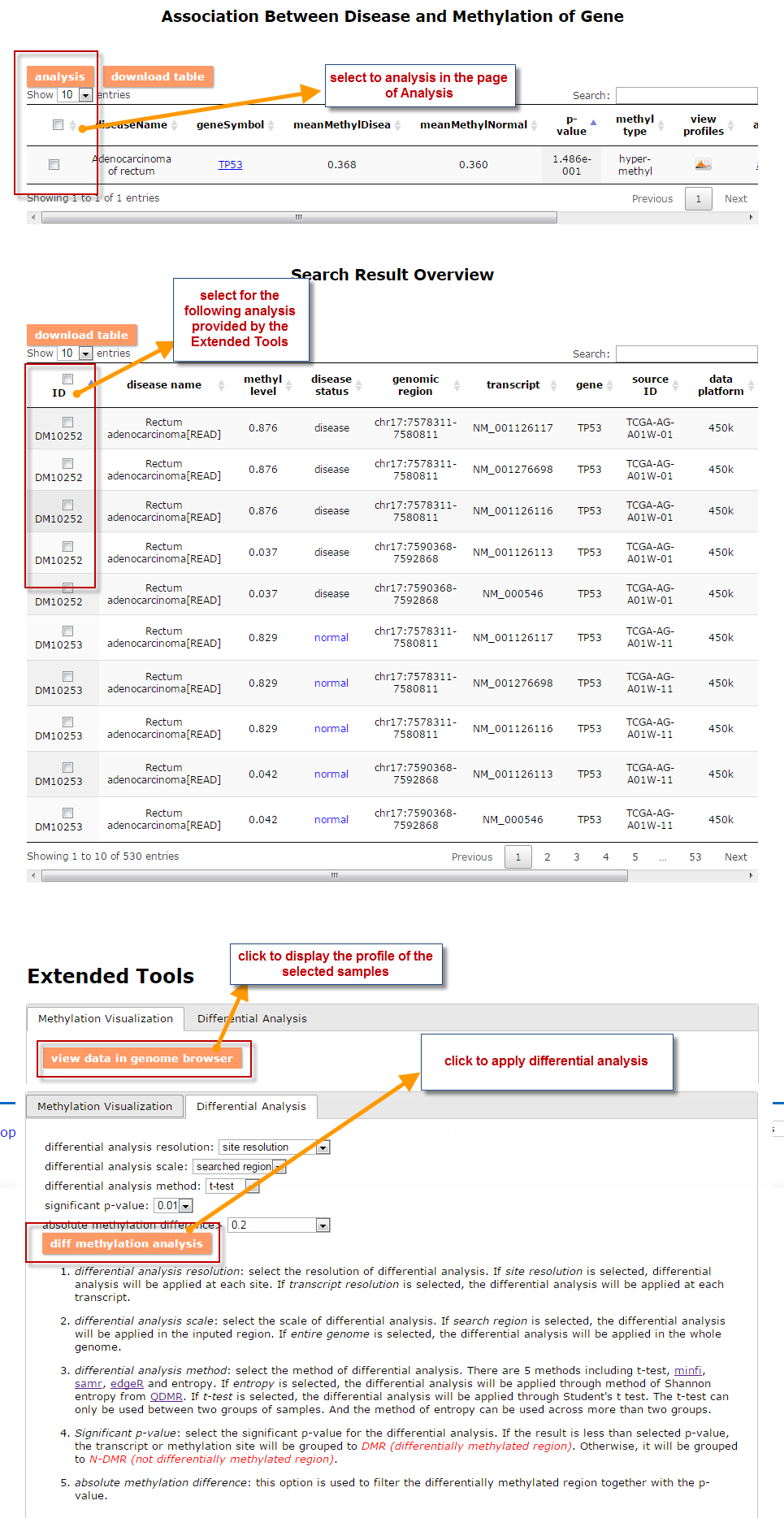
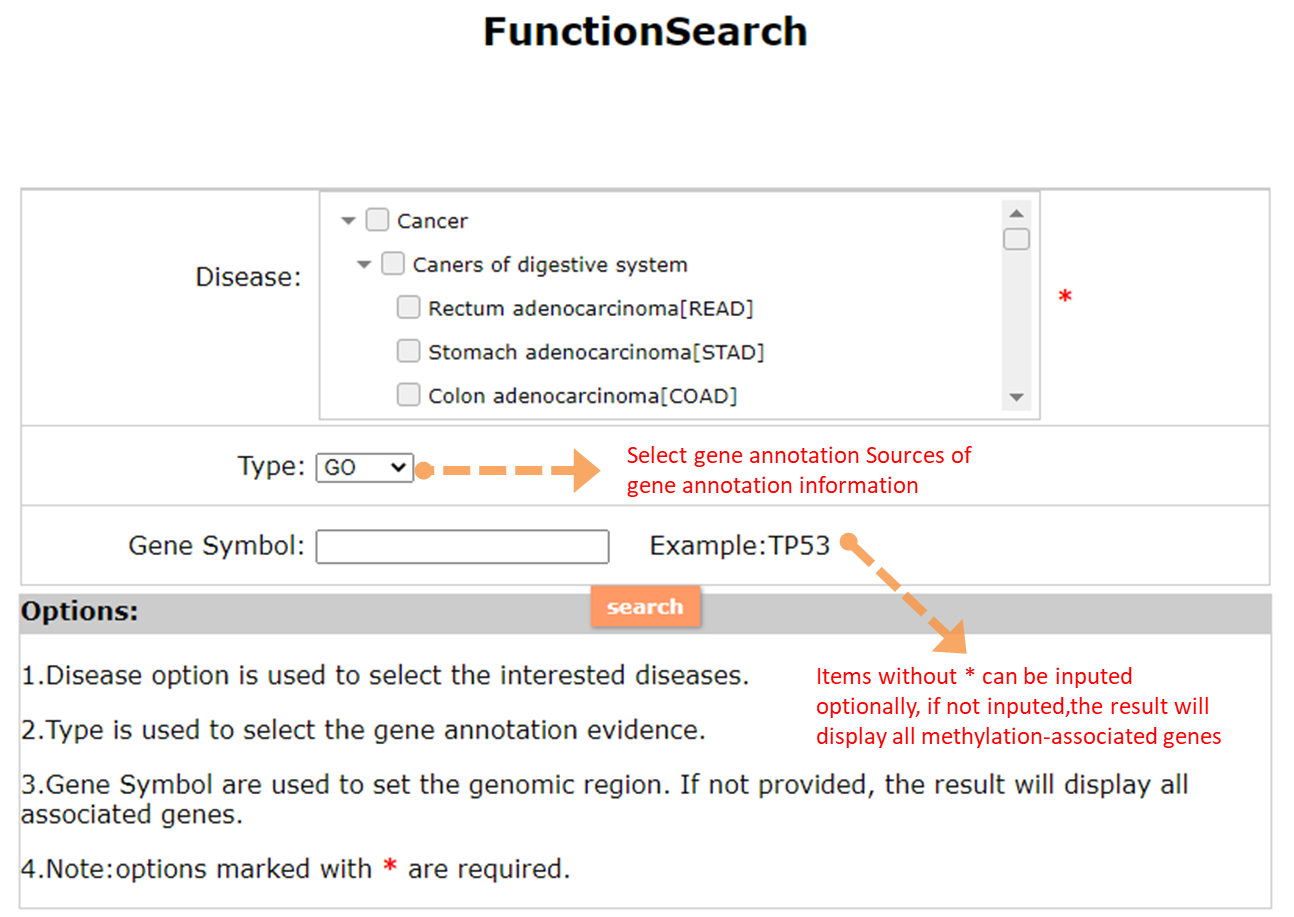
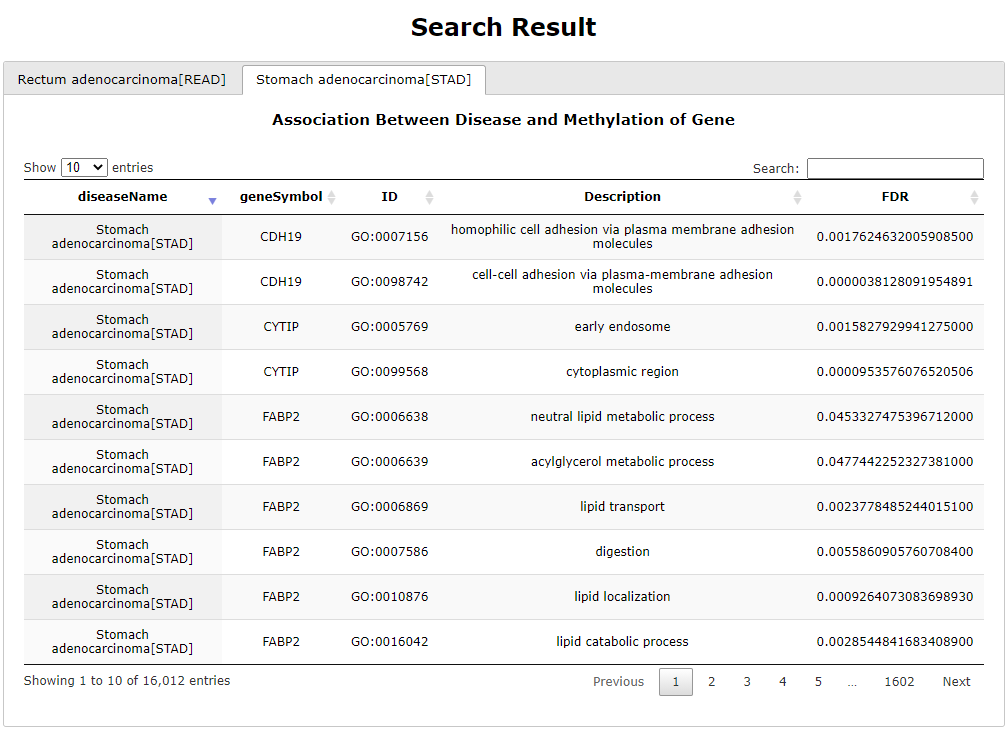
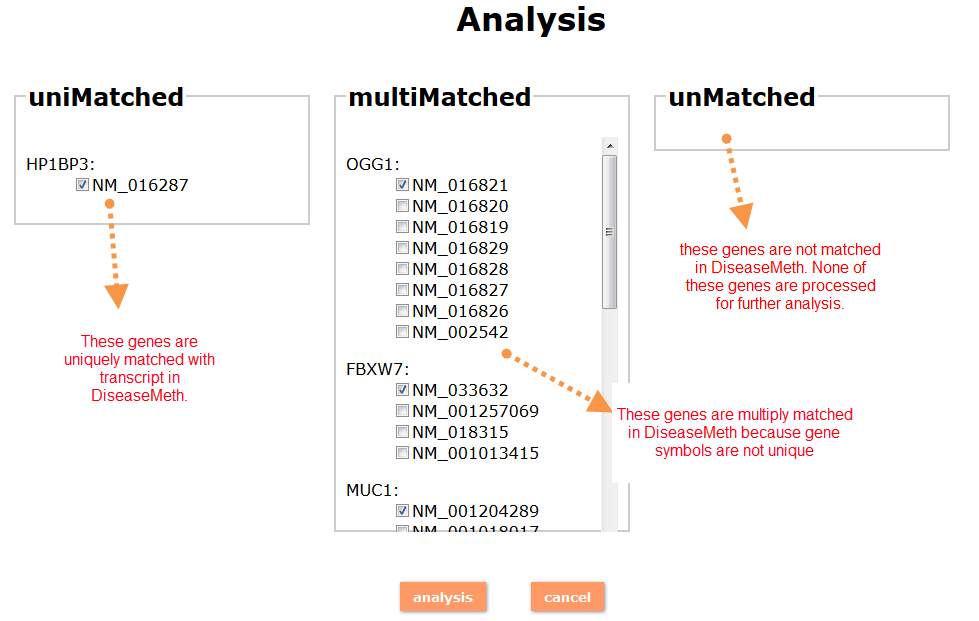
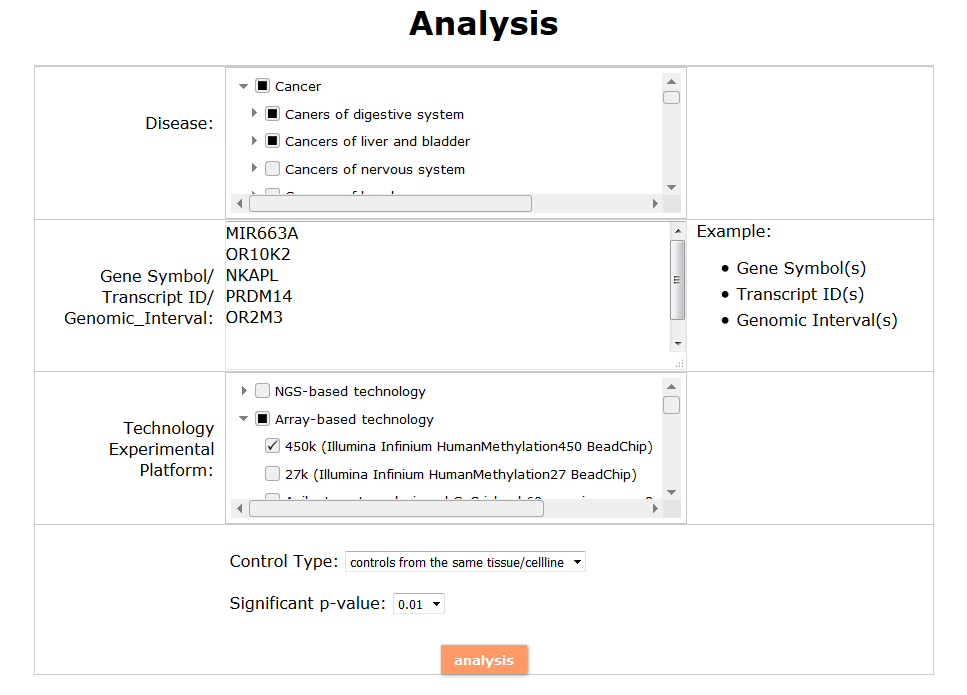
(1) The Search Engine was redesigned into three query entrance: DisMethSearch (query disease methylome or DNA methylation level for a specified gene/genomic position), DiseaseSearch (query DNA methylation level for disease associated genes by disease type) and DiseaseGeneSearch (query DNA methylation level across diseases for a specified gene/genomic position),GeneFunctionSearch (query The biological function of genes across diseases for a specified gene/genomic position)
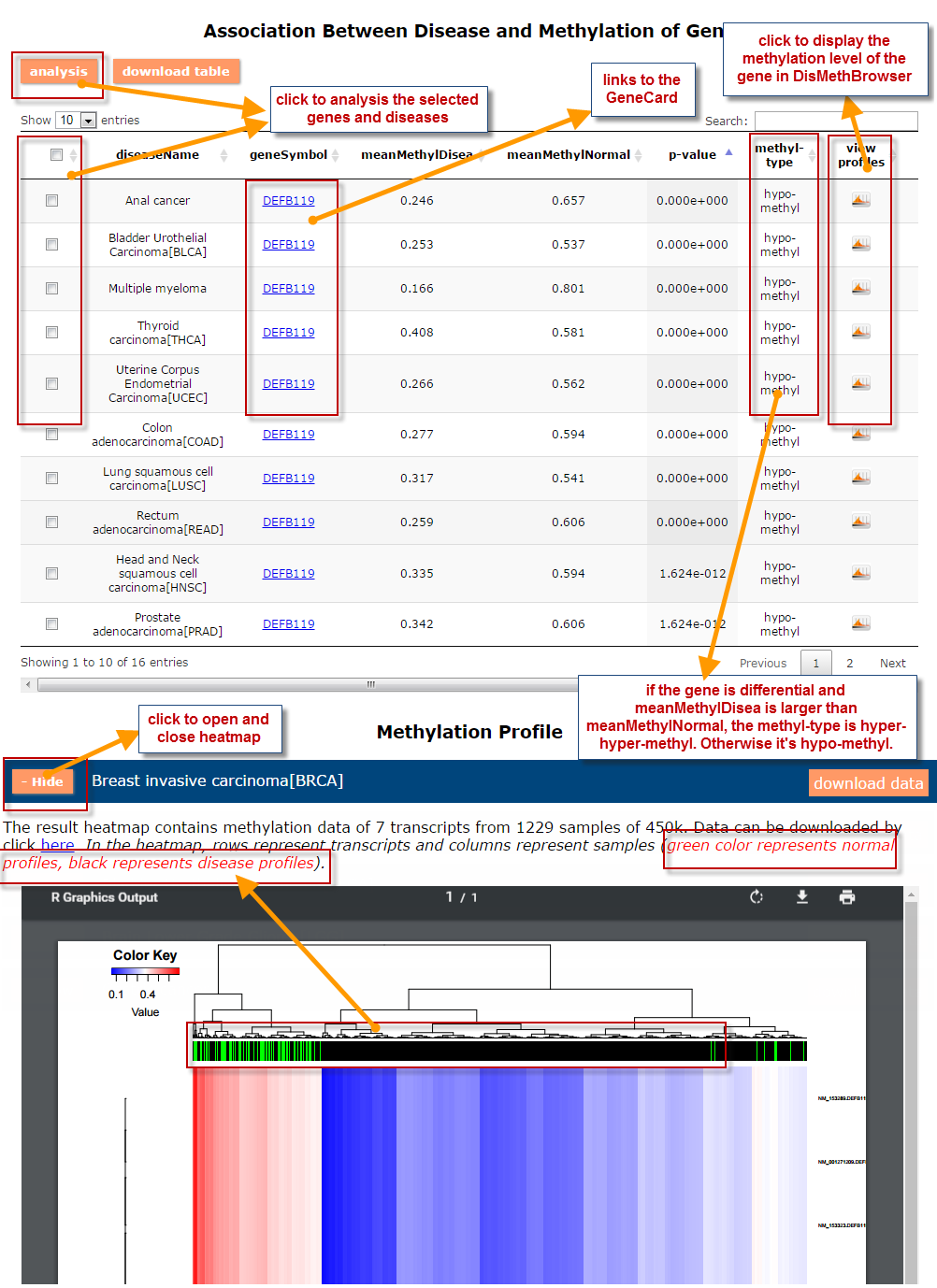
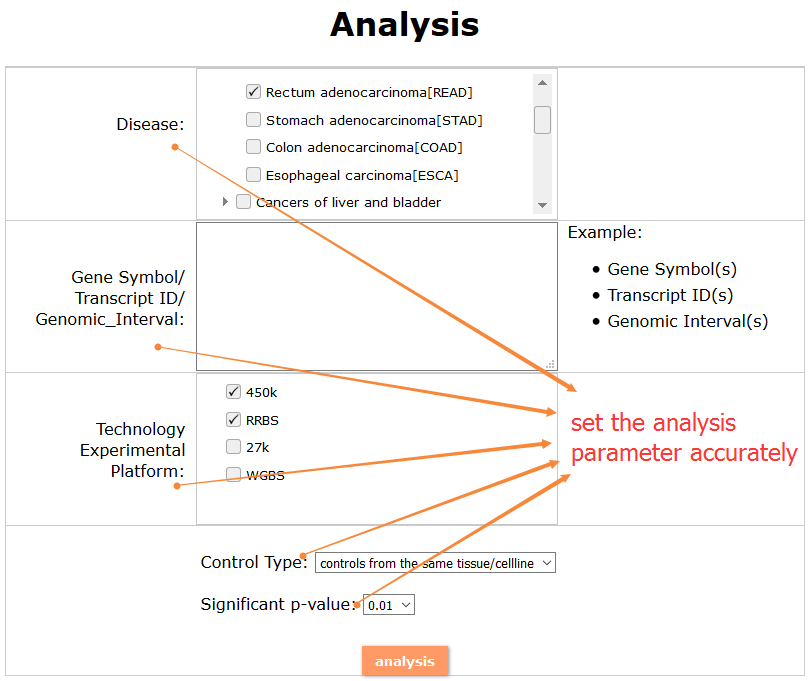
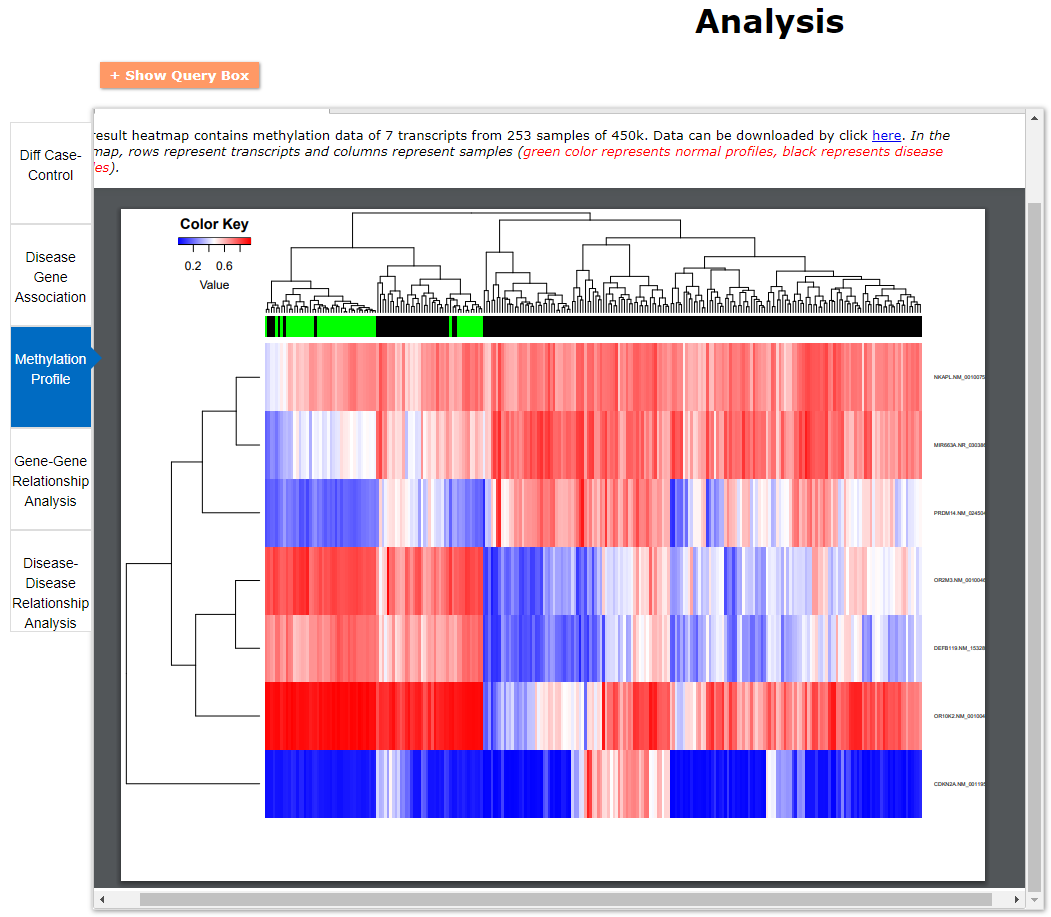
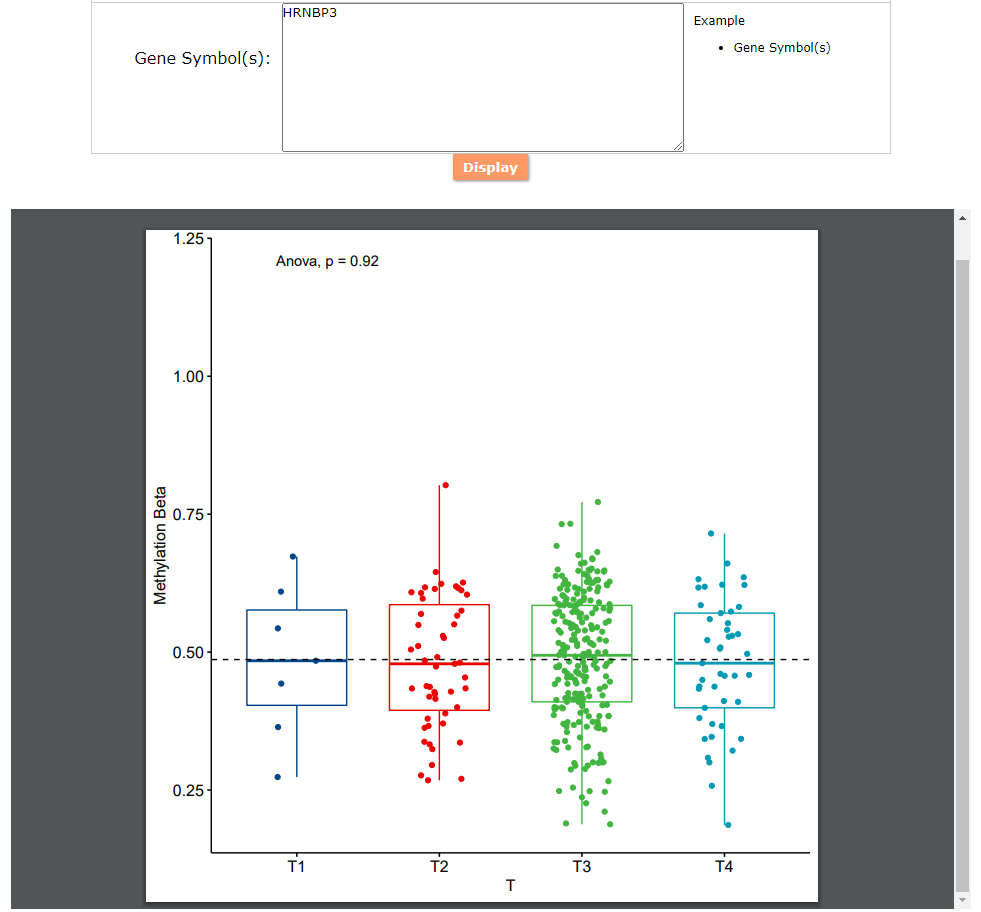
(2) For exploring the biological significance of DNA methylation across diseases or case-control samples through large amount of data, DiseaseMeth version 3.0 contains three new tools for the genetic analysis: cluster analysis (clustering the methylation profile of selected diseases samples within input genes and providing a heatmap view of the result to users), functional annotation (annotate the function of selected genes and obtain the functional cluster of these genes) and survival analysis (used to judge if a gene is related to the survival time of patients with a specific cancer).
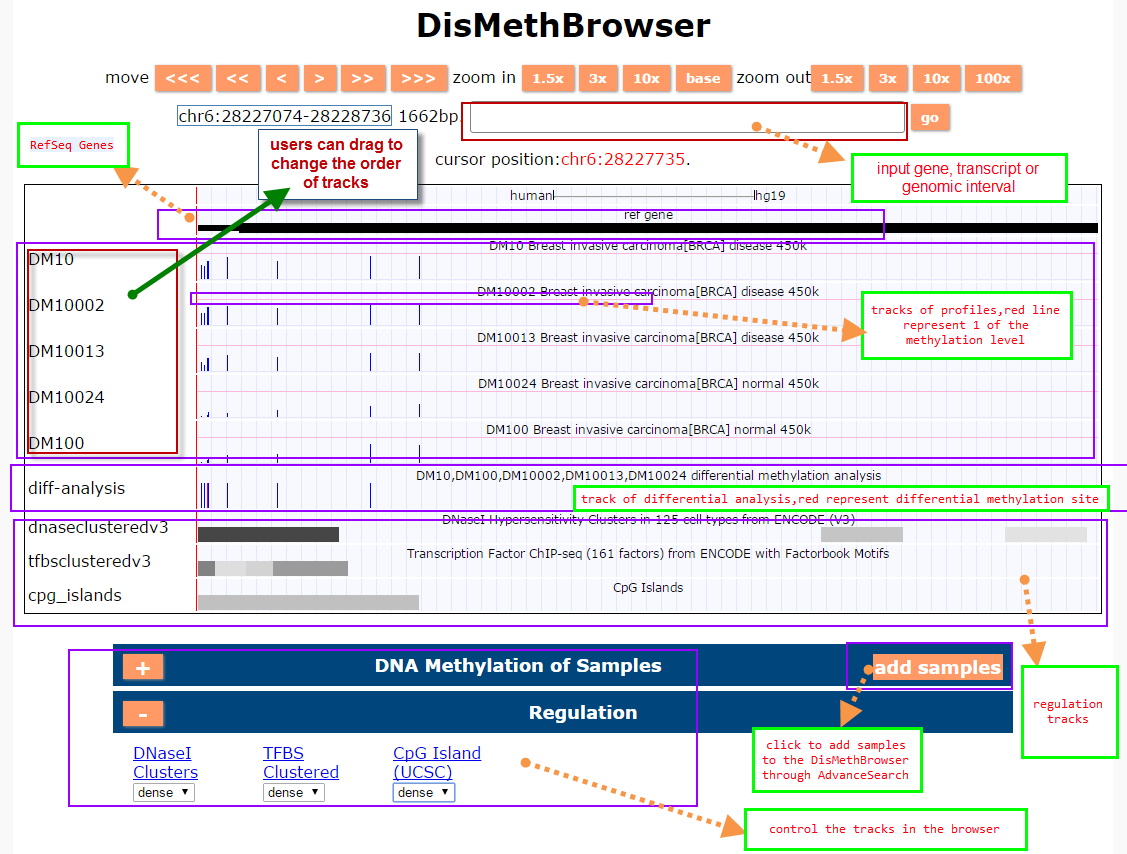
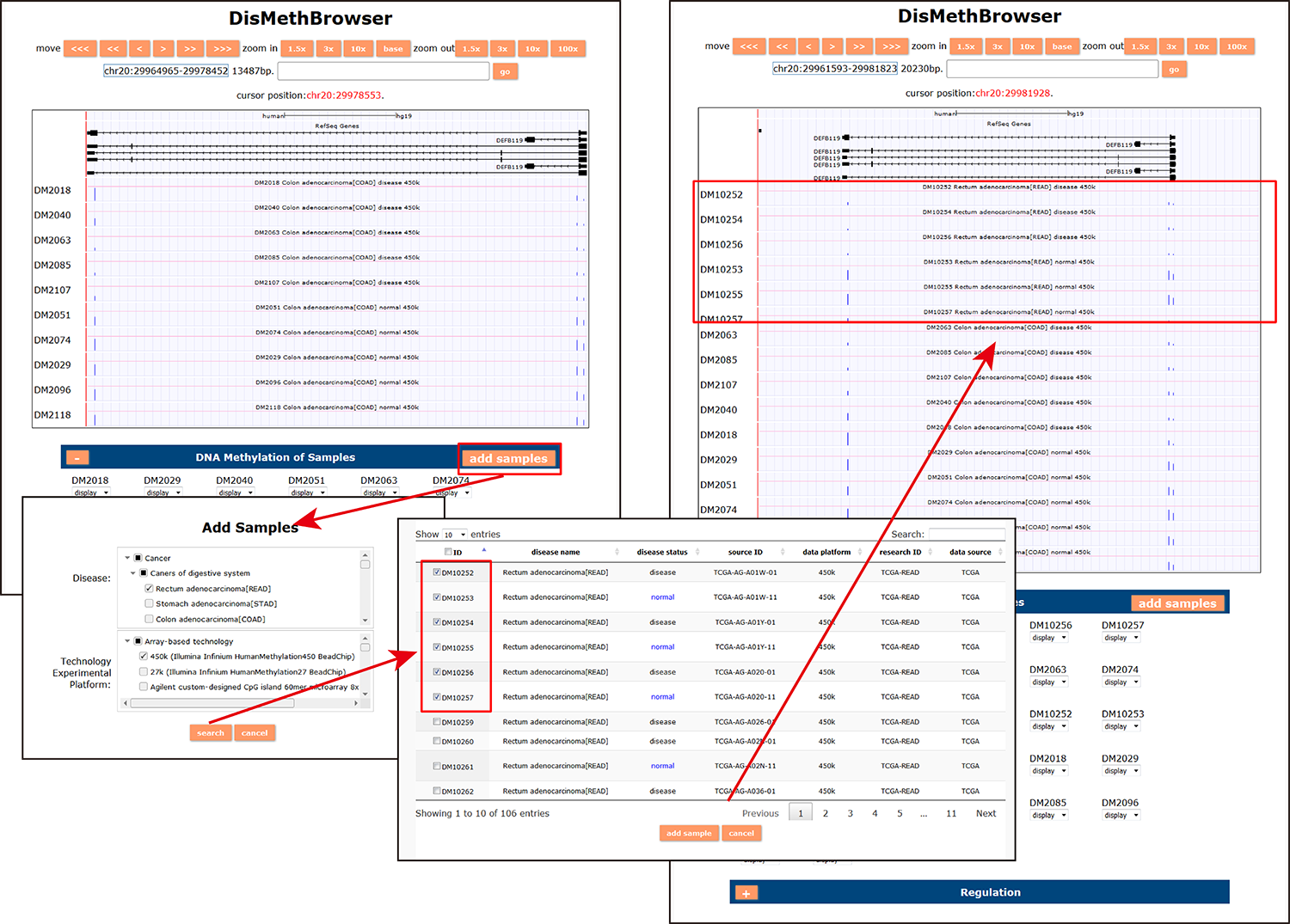
(3) DisMethBrowser: A newly developed disease DNA methylation visualization tool, by showing disease associated differential methylation status, methylation level for different samples, ref-genome transcripts and transcriptional regulators in independent tracks.
-
the orange trace
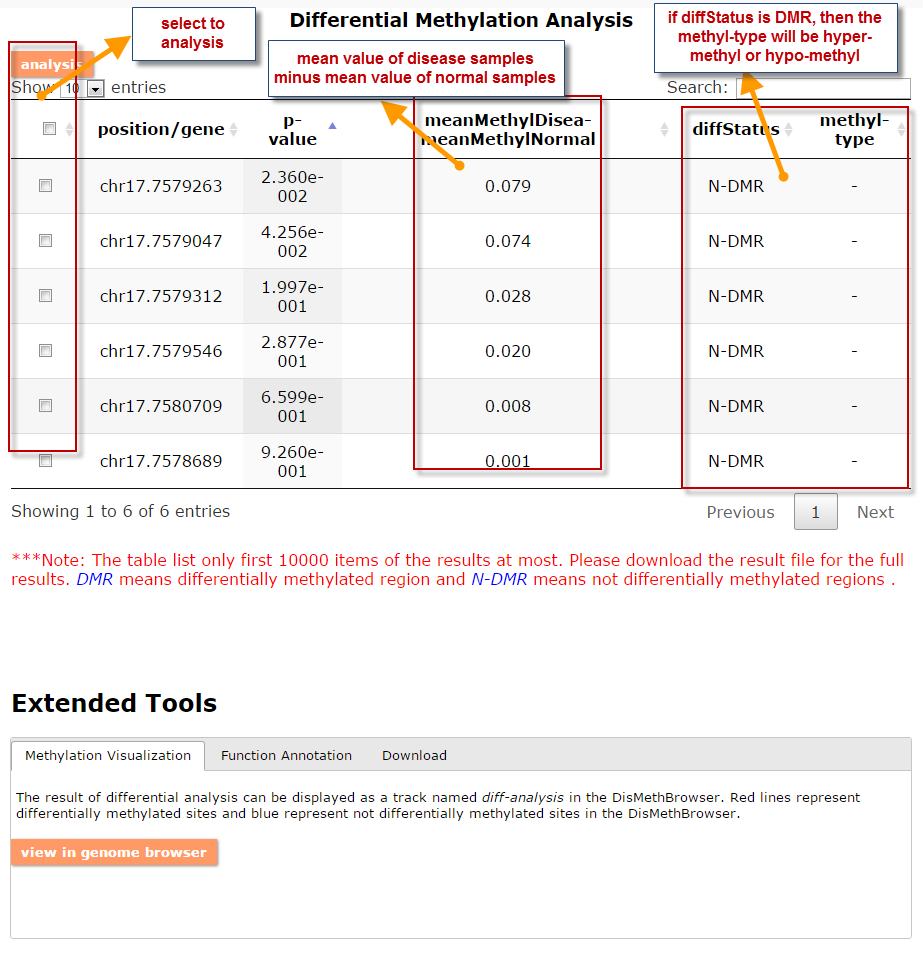
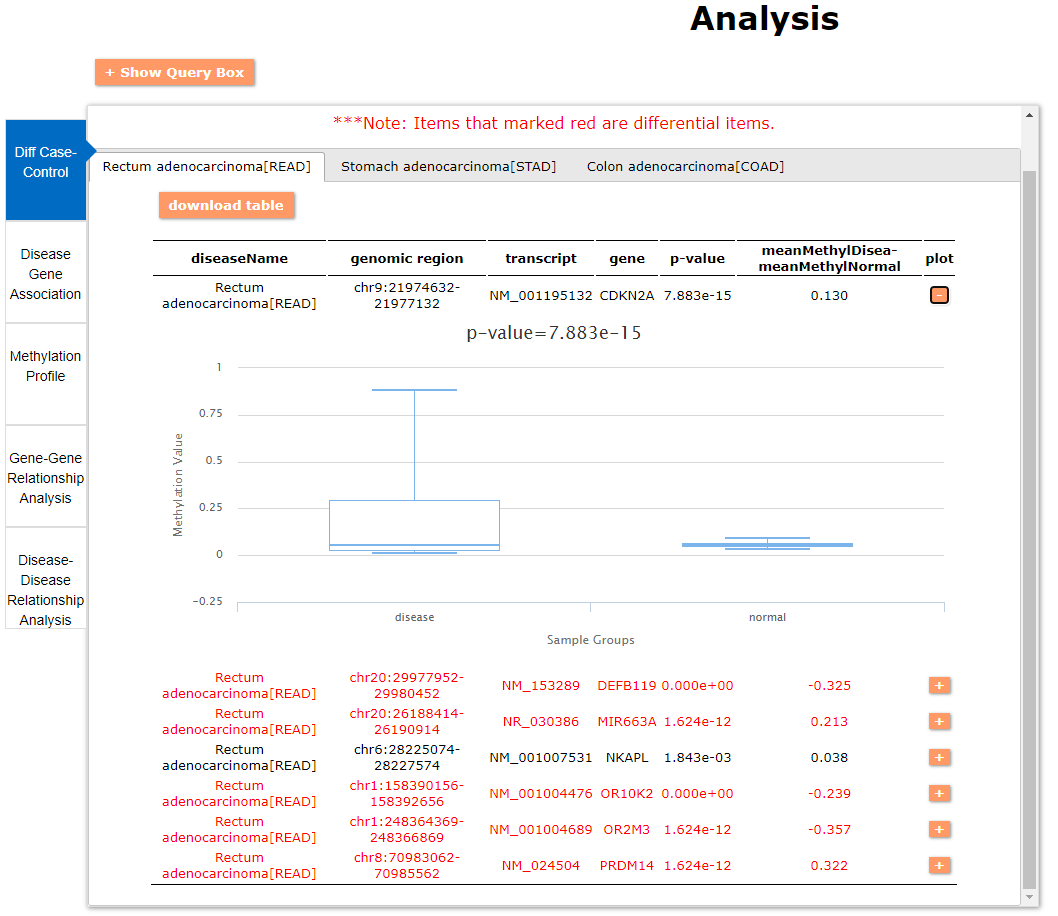
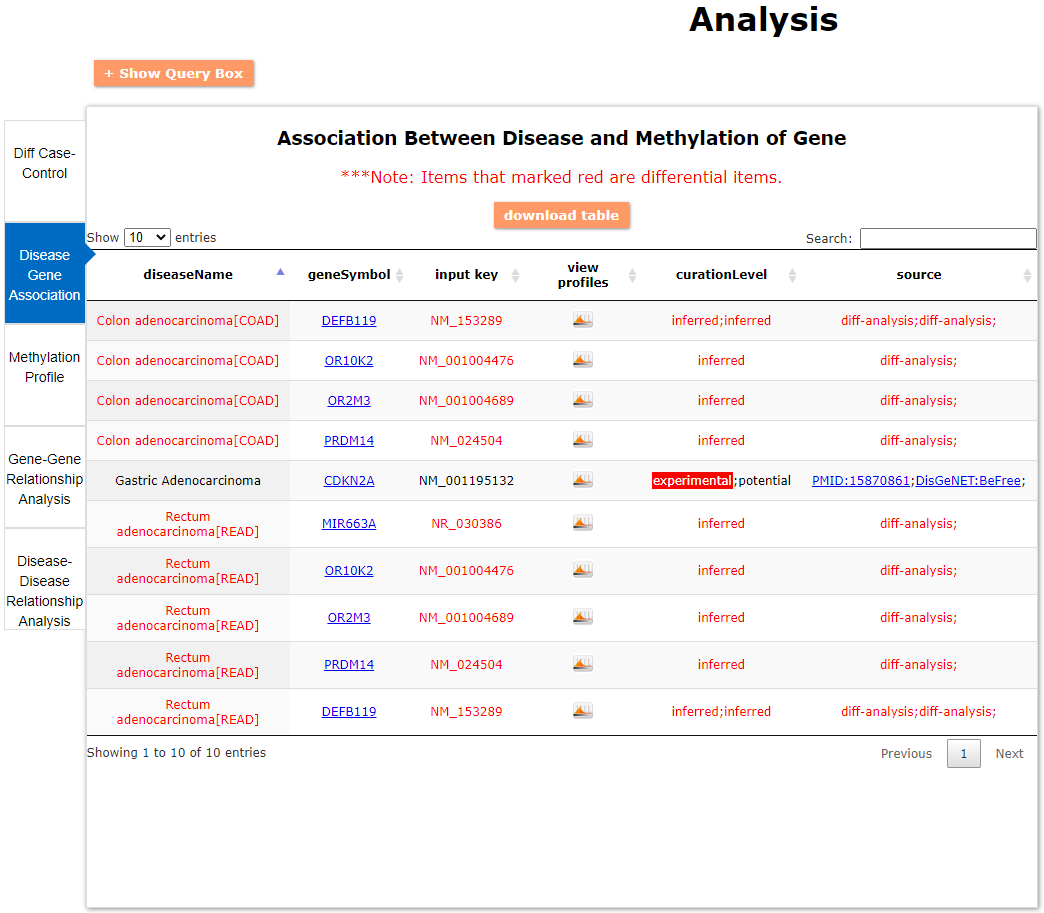
(1) The differential DNA methylation analysis tool is updated by integrating the method of Student's t-test and Shannon entropy to analyze the difference across two groups or more than two groups. And the significantly differential methylated genes are defined as the disease associated genes.
(2) Tools: A newly developed cancer prognosis and co-methylation visualization tool, by showing caner associated differential methylation level for different samples in differential clinical factors.
-
-
Extended Datasets in DiseaseMeth v3.0
In DiseaseMeth version 3.0, the number of disease types was increased from 88 to 162. We screened 4,708 methylation datasets in the GEO and TCGA database from Oct 1, 2015 to Jan 31, 2021, collecting a total of 247 high-throughput datasets related to human diseases from ChIP and 29 sequencing datasets. Total number of samples were increased from 32,701 to 49,949. Moreover, the associations between disease and gene from experiment were increased from 679,602 to 1,485,099 by manual literature search from Pubmed.